How to measure brain activity in animals
Unlike measuring brain activity in humans, the techniques used to record brain activity in animals provide much greater detail than techniques designed for human use, working on the scale of 1–1000 neurons rather than millions or billions of neurons. The result is that the questions addressed in human neuroscience research are fundamentally different from those posed in animal research. Human studies are largely about localisation of function and how behavioural performance in a task correlates with gross level brain activity. In contrast, animal research is often aimed at understanding how neurons interact with each other, to encode and process information.
Implanted Multi-electrode Arrays (MEAs)
Whereas EEG uses electrodes on the scalp surface, and ECoG electrodes sit on the brain surface, some electrode arrays are inserted into the brain itself. As a broad class, these inserted electrodes are called multi-electrode arrays (MEAs). Because they are inserted into the brain, the 10s or 100s of electrodes end up very close to, but outside of, individual neurons, where they can record action potentials (spikes).
Furthermore, based on their size and shape, the spikes can be assigned as coming from different neurons. With ~100 electrodes on an individual device, MEAs can track ~10–1000 neurons over days, months or even years, providing a direct readout of how a small neuronal circuit operates over long periods of time. This is potentially powerful information.
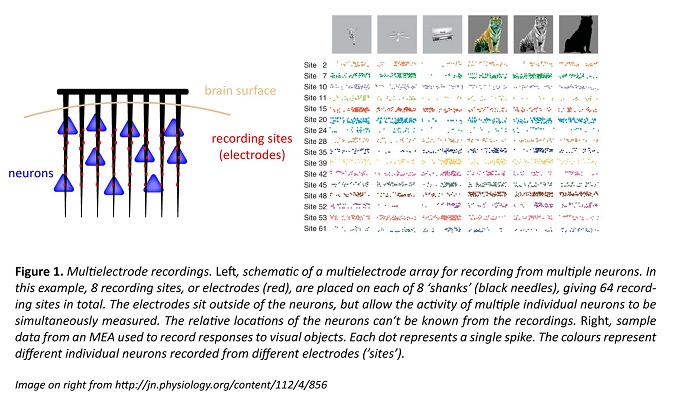
MEAs provide excellent information on when particular neurons spike. However, it is difficult to determine what type of neuron is giving the signal; even the fundamental distinction between excitatory and inhibitory neurons can’t always be made reliably, and there are tens if not hundreds of different neuron types in the brain. If we can’t tell what type of neuron is responsible for a given signal, we can’t figure out how the circuit really behaves. A solution lies in a more recent technique, fluorescence imaging of neuronal activity.
Fluorescence Imaging of Neuronal Activity
Researchers can use fluorescent ‘indicators’ to optically record neuronal activity in roughly the same number of neurons as an MEA can record (Fig. 2). Also like MEAs, many of these fluorescent indicators allow the activity of multiple individual neurons to be tracked, and this can take place in a behaving animal, as shown in the video below. In this video, the small flashing spots represent single neurons.
The original fluorescent indicators were organic dyes injected into the brain, but they didn’t specifically report the activity for a particular type of neuron. More recently, genetic advances have enabled specific types of neurons to be the only ones labelled, meaning that researchers can easily focus their attention on the activity of genetically defined neuronal populations. This is the primary advantage of fluorescence imaging over MEAs.
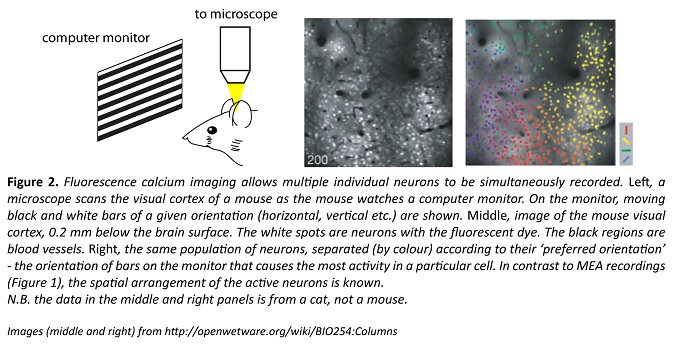
Currently, most fluorescent indicators allow neuronal activity to be distinguished on a timescale of tens of milliseconds; much more precise than for fMRI (and with single-cell resolution) but still inferior to electrical recordings. Because it is an optical technique, fluorescence imaging in animals has two significant limitations – the skull must be removed or thinned, and structures deep within the brain are inaccessible to the light even when the skull is absent.
Nevertheless, fluorescence imaging of neuronal population activity is a rapidly growing and extremely promising approach for understanding how different types of neurons interact with each other. It’s likely that in the next 10 years, the temporal resolution will rival electrical recordings and that imaging much deeper in animal brains will be more feasible.
Patch-clamp recordings
Patch-clamp recordings are the most precise. An fMRI can record whole brain activity at slow speeds, EEG and ECoG can record mass activity across the brain’s outer surface (cortex) at high speeds, and MEAs and fluorescence imaging allow the spiking of hundreds of individual neurons to be followed at very high or moderately high speeds, respectively.
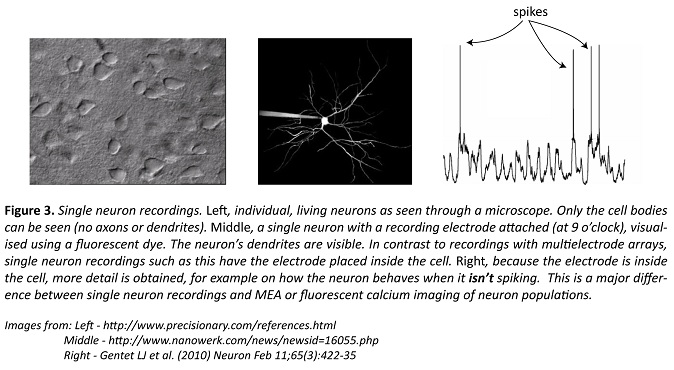
Single-cell patch-clamp recordings (the technique garnered a Nobel Prize in 1991) are made by placing an electrode inside the neuron, a feature that distinguishes them from all other techniques described so far, including EEG or even MEAs. This allows the researcher to record what the neuron is doing when it’s not spiking.
When combined with tricks to manipulate the recorded call, information can be gathered on what inputs the neuron is receiving. Neither MEAs nor fluorescence imaging provide such direct information on a neuron’s inputs, and so despite the limitation of only recording one of the brain’s billions of neurons, single-cell recordings can still provide extremely valuable, detailed information. Still, it’s near impossible to know where the inputs are coming from, and making single-cell recordings in behaving animals is extremely difficult.
Summary
There are many different ways to record brain activity, a necessity that arises in part because of the vast physical and temporal scales at which the brain operates. The question to be answered dictates the technique(s) that should be used. To get detailed information on how neurons interact and how small circuits process information, invasive tools are used in animals; we can’t study individual neurons non-invasively. And to know where in the human brain neural activity is localised for particular functions, researchers use non-invasive approaches that allow cognitive performance to be correlated with coarse-level brain activity.
QBI researchers using multielectrode recordings
Professor Pankaj Sah
Associate Professor Bruno van Swinderen
QBI researchers using fluorescence imaging (to record neuronal activity)
Professor Stephen Williams
Professor Pankaj Sah
Professor Joe Lynch
Professor Geoff Goodhill
QBI researchers using patch-clamp recordings
Professor Stephen Williams
Professor Pankaj Sah
Professor Joe Lynch
Associate Professor Bruno van Swinderen
